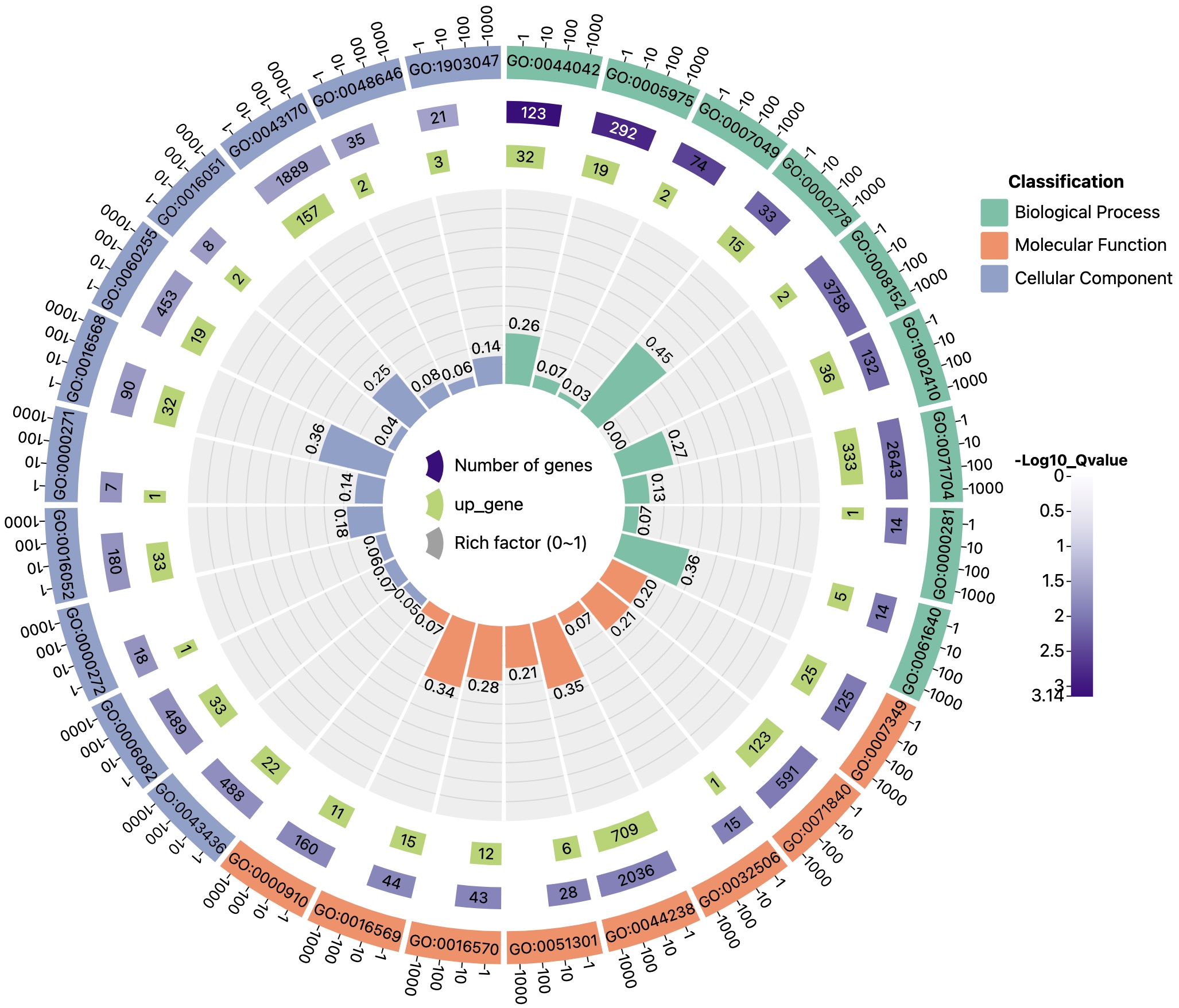
data file format
The data file can be in one of supported formats (xlsx, xls, tsv or csv), containing 5 or 6 columns.
Column 1: The ID information of GO term.
Column 2: The classification name corresponding to this ID (Provide the standard classification names of databases such as GO/KEGG pathway, and note that the classification names are in capital letters). The functional legends in the figure are displayed by classification according to the information in this column.
Column 3: P-value/Q-value, plot based on P-value or Q-value, and enter only one of them.
Column 4: The total number of genes identified in this classification, also called background genes.
Column 5 and column 6: There are two formats for input. When up and down regulated genes are distinguished, the number of up-regulated and down regulated genes is input in column 5 and column 6 respectively. When up-down regulation is not distinguished, only the total number of differential genes in column 5 needs to be entered.
Example file 1 (6 columns in total, data table for distinguishing up and down regulated genes):
| ID | classification | pvalue | all | up_gene | down_gene |
|---|---|---|---|---|---|
| GO:0044042 | Biological Process | 0.000710215 | 123 | 32 | 32 |
| GO:0005975 | Biological Process | 0.001451882 | 249 | 19 | 19 |
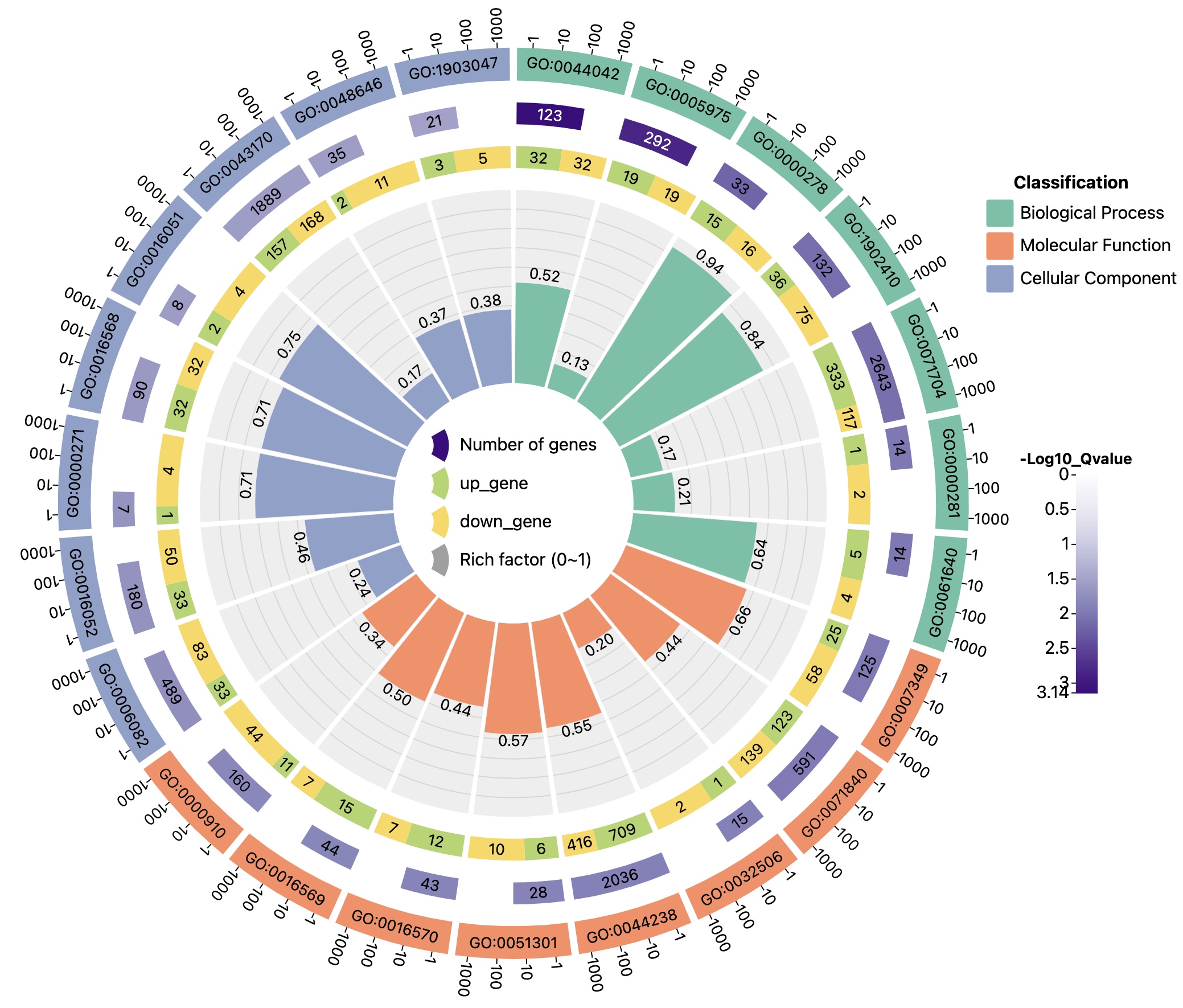
Example file 2 (5 columns in total, no distinction between up and down regulated genes)
| ID | classification | pvalue | all | differ gene |
|---|---|---|---|---|
| GO:0044042 | Biological Process | 0.000710215 | 123 | 32 |
| GO:0005975 | Biological Process | 0.001451882 | 249 | 19 |